GEOMI is a network visualisation and analysis platform originally developed at National ICT Australia (NICTA). The program and its source code are released under the LGPL licence. For more information, please click here.
The SBI version of GEOMI enables visualisation of protein-protein interaction data in four-dimensions, and mapped with proteomic data as described in:
- Li SS, Xu K, Wilkins MR (2011). Visualisation and analysis of the complexome network of Saccharomyces cerevisiae. Journal of Proteome Research, 10: 4744-56. [abstract]
- Goel A, Li SS, Wilkins MR (2010). Four-Dimensional Visualisation and Analysis of Protein-Protein Interaction Networks. Proteomics, 11: 2672-82. [abstract]
- Ho E, Webber R, Wilkins MR (2008). Interactive three-dimensional visualization and contextual analysis of protein interaction networks. Journal of Proteome Research, 7: 104-12. [abstract]
Download GEOMI (SBI version) [zip]
Requires Java and at least Java 3D 1.5.
GEOMI User Guide [pdf]
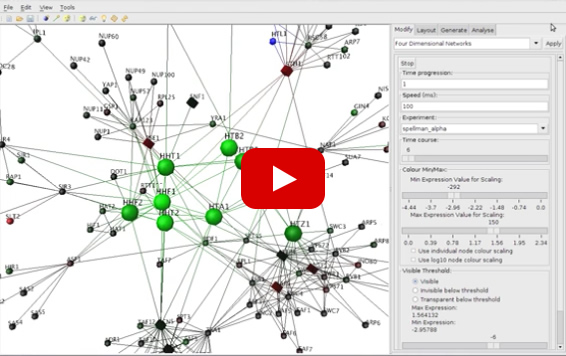
4-D Visualisation of Protein-Protein Interaction Networks
More videos can be viewed on our Youtube channel.
DNA mismatch repair proteins
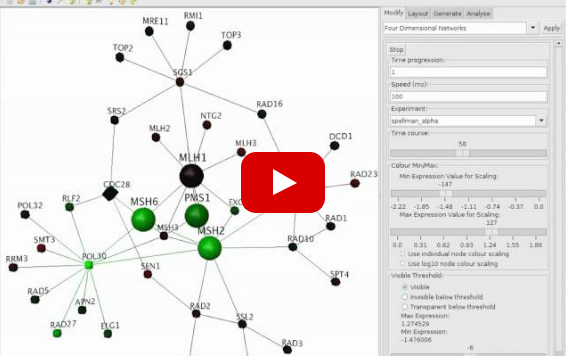 Cell cycle gene expression data has been mapped onto DNA mismatch repair proteins MLH1, MSH2, MSH6, PMS1 and nearby interacting proteins in yeast.
Green: down regulation,
Red: up regulation,
Black: no change in expression.
Colour intensity indicates level of expression change.
Cell cycle gene expression data has been mapped onto DNA mismatch repair proteins MLH1, MSH2, MSH6, PMS1 and nearby interacting proteins in yeast.
Green: down regulation,
Red: up regulation,
Black: no change in expression.
Colour intensity indicates level of expression change.
XWG Files
The XWG (XML Wilma Graph) file format is used by GEOMI to visualise networks. To view these files, move the downloaded files to the data directory in GEOMI. Make sure the directory also contains the WilmaGraph.dtd file.
| YEAST (SACCHAROMYCES CEREVISIAE) REFERENCES | |
|---|---|
| CMnetwork.xwg | complexome network of Gavin et al. (2006) dataset [abstract] |
| CMnetwork_GOprocess.xwg | complexome network, with mapped putative function |
| 4D_Histones.xwg | 4D network of histone proteins |
| 4D_MLC1.xwg | 4D network of hub protein MLC1 |
| 4D_DNAMismatchRepair.xwg | 4D network of DNA mismatch repair proteins |
| 4D_YPT52.xwg | 4D network of hub protein YPT52 |
| 4D_CentralCarbon.xwg | 4D network of central carbon network metabolism proteins |
| all.xwg | interactome of Bertin et al. (2007) dataset [abstract] |
| all_bcc.xwg | biggest connected component of the above |
| cytoplasm.xwg | interactome within the cytoplasm |
| cytoplasm_bcc.xwg | biggest connected component of the cytoplasmic interactome |
| mitochondria.xwg | interactome within the mitochondria |
| nucleus.xwg | interactome within the nucleus |
| nucleus_bcc.xwg | biggest connected component of the nuclear interactome |
| HUMAN (HOMO SAPIENS) REFERENCES | |
|---|---|
| cell_membrane_and_secreted.xwg | interactome of cell membrane and secreted proteins |
| cytoskeleton.xwg | interactome of cytoskeleton proteins |
| ER_to_Golgi.xwg | interactome of ER and Golgi proteins |
| mitochondria.xwg | interactome of mitochondria proteins |
| HELICOBACTER PYLORI REFERENCES | |
|---|---|
| hpylori_all_annotated.xwg | interactome of Rain et al. (2001) dataset [abstract] |
| ESCHERICHIA COLI | |
|---|---|
| E_coli_protein_interaction_networks_database.zip | E. coli protein interaction networks database |
| ecoli_all.xwg | interactome of Hu et al. (2009) dataset [abstract] |