Proteomic-Genomic Nexus
The Proteomic-Genomic Nexus (PG Nexus) is a Java-based software package that is designed to integrate genomic and transcriptomic data generated from next-generation sequencing with proteomic data generated from protein mass spectrometry. The tools were built by Intersect Australiaas part of an ANDS-funded project within the Applications Programme.
By using the PG Nexus, users will be able to:
- Co-visualise genomics, transcriptomic and proteomic data using the Integrated Genomics Viewer(IGV).
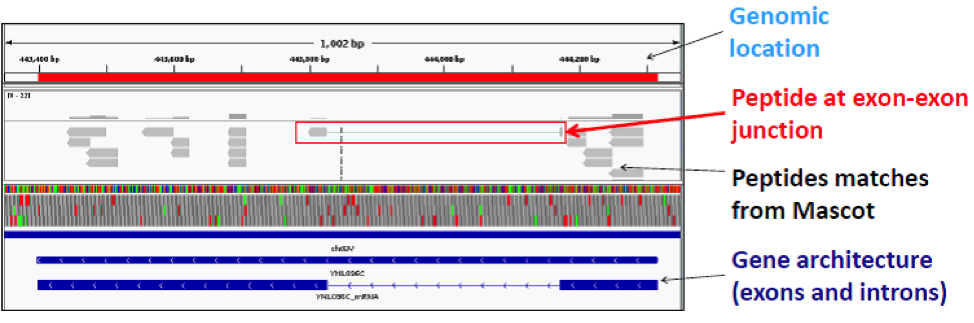
- Validate the existence of genes and mRNAs using peptides identified from mass spectrometry experiments.
- Validate alternatively spliced mRNA isoforms by searching for peptides that span across exon-exon junctions.
Download the PG Nexus via GitHub.
For more information, please refer to the article:
Chi Nam Ignatius Pang, Aidan P. Tay, Carlos Aya, Natalie A. Twine, Linda Harkness, Gene Hart-Smith, Samantha Z. Chia, Zhiliang Chen, Nandan P. Deshpande, Nadeem O. Kaakoush, Hazel M. Mitchell, Moustapha Kassem, and Marc R. Wilkins (2014). ). Tools to Covisualize and Coanalyze Proteomic Data with Genomes and Transcriptomes: Validation of Genes and Alternative mRNA Splicing. Journal of Proteome Research, 1: 84-98. [abstract]
Alternatively, click on the video below or visit our wiki page here!
Introduction to the PG Nexus:

Downloadable Files
- Database and input files for the PG Nexus can be downloaded here.These files were used to validate protein isoforms with RNA-Seq data.
- The human genome (version Hg19) downloaded here. It is formatted with one chromosome per FASTA file and is compatible with the pipeline.
- The human genome (version 75) can be downloaded here. These files were used to validate protein isoforms with RNA-Seq data.

